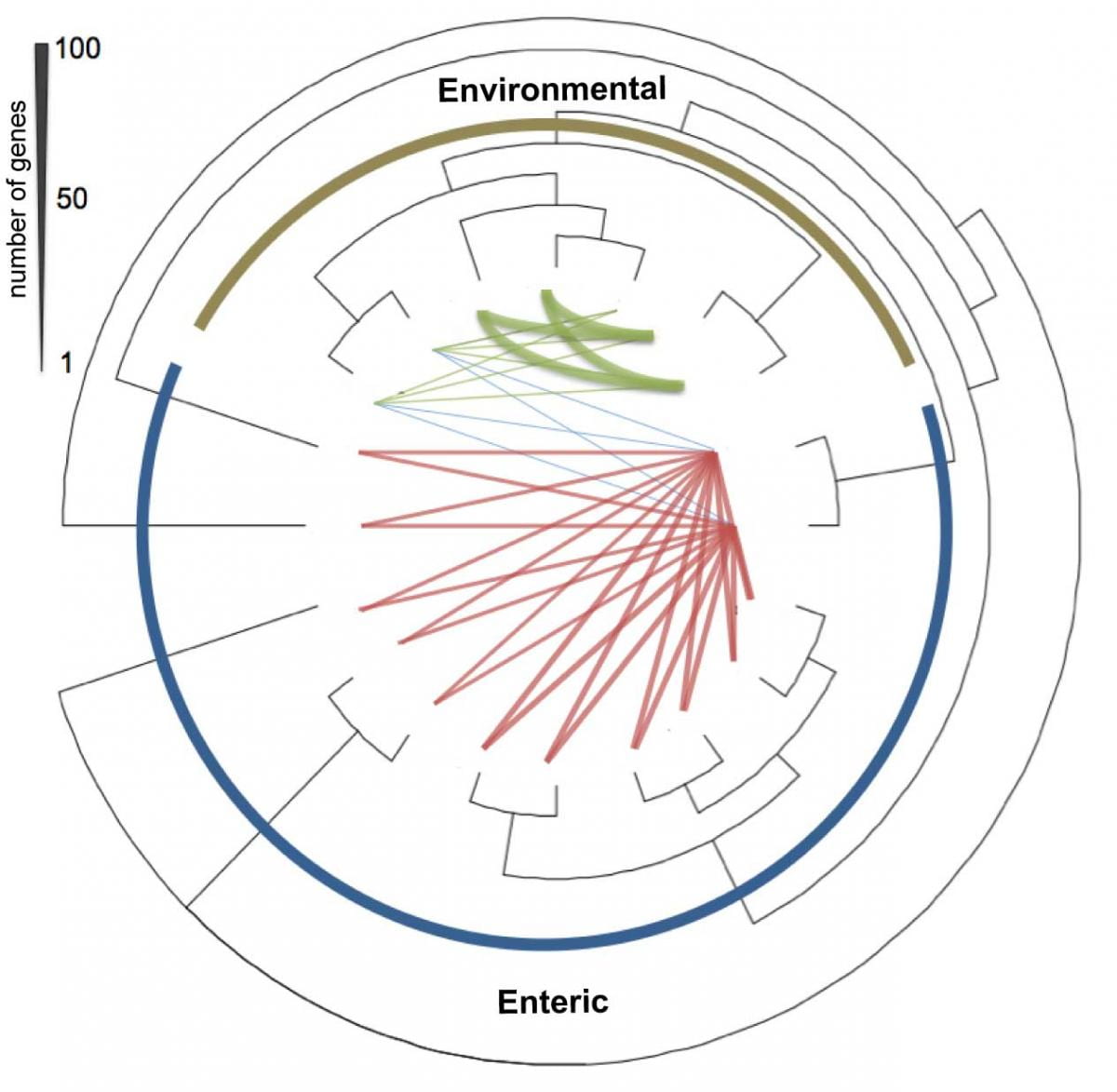
1. Comparative genomics to better understand microbial speciation and evolution
We perform comparative analyses of genomes of isolates to evaluate how bacteria adapt to environmental perturbations and what is the relative importance of the genetic mechanisms of genome evolution and plasticity. This work has also important implications for the bacterial species definition. We study several important groups including, but not limited to, E. coli, Campylobacter jejuni, Shewanella spp, Burkholderia spp, and Synechococcus spp. For instance, see our PNAS paper on E. coli:
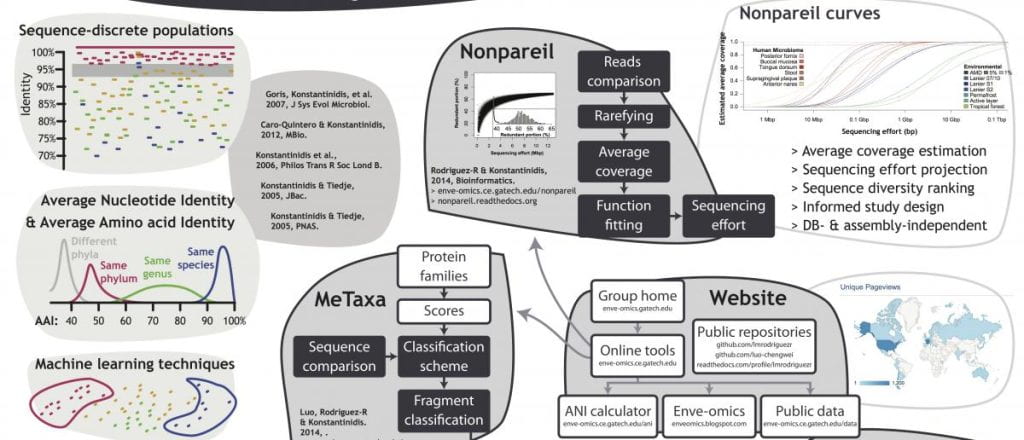
2. Bioinformatics tools for microbial genomes and metagenomes
With support from the NSF “Advances in Biological Informatics” program, we are developing novel bioinformatics tools for specialized tasks in the analysis of microbial genomes and metagenomes such as how to determine the level of the coverage of a microbial community obtained by sequencing (Nonpareil), how to detect metagenomic reads encoding a target gene of interest (ROCker), an ANI/AAI calculator, and a webserver to taxonomically identify genome sequences using the ANI/AAI approach (a.k.a. the “genome-equivalent” of the RDP/Silva 16S rRNA gene databases). For further details, see our “Tools” webpage from the menu above and the link below:
http://nsf.gov/awardsearch/showAward?AWD_ID=1356288
The Microbial Genome Atlas (MiGA) webserver for taxonomic identification of complete or partial genome sequences is available here:
And the link to the new NSF ABI grant that supports MiGA’s expansion to include the uncultivated microbes can be found here:
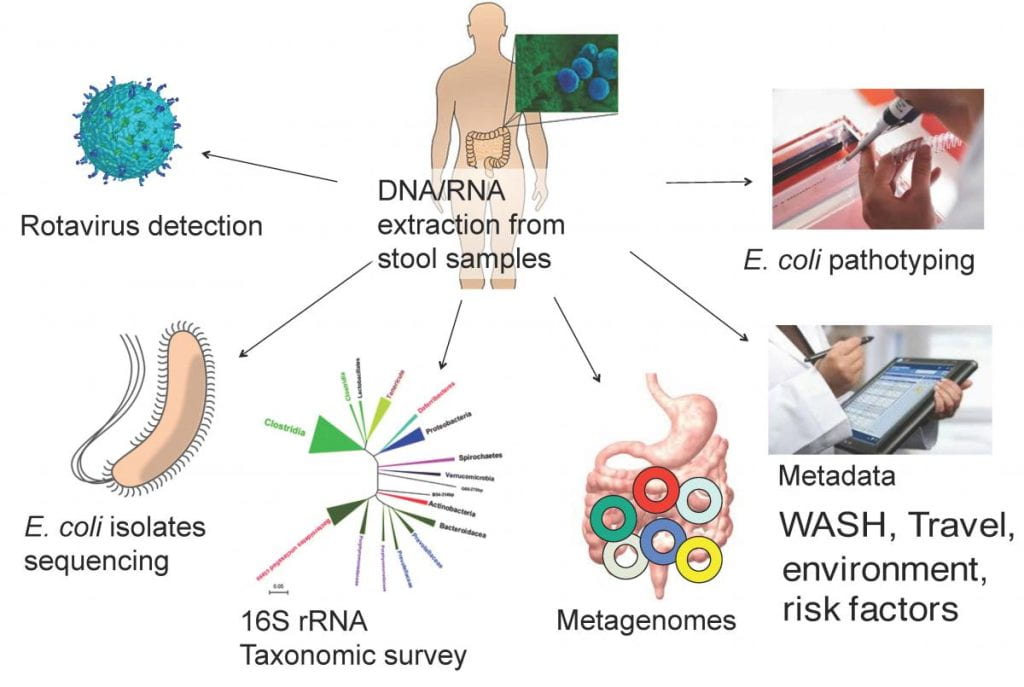
3.Metagenomic-based diagnostics of enteric pathogens and analysis of human-associated microbial communities
With support from NIH and together with collaborators at Emory University, CDC and elsewhere, we examine the impact of sanitation interventions (WASH) on enteric pathogen infection of young children in developing communities in South Africa. Another NIH-supported project is to understand the importance of gut microbiome diversity in resisting or recovering from enteric infections in a rural-to-urban gradient in South America, and develop culture-independent, metagenomic methods for detecting the etiological agents of infectious diarrhea. For further details, see:
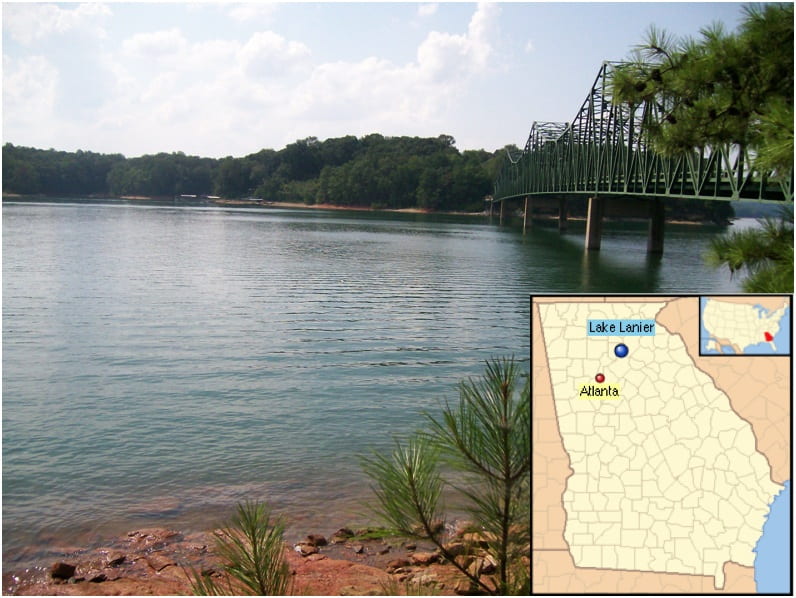
4.Time-series metagenomics of Lake Lanier, Atlanta, GA
Lake Lanier is an important freshwater lake for the South East US as it represents the main source of drinking water for the Atlanta metropolitan area and is extremely popular with boaters, houseboats, jet skiers and others. To better characterize the microbial communities associated with human-impacted freshwater ecosystems, we have begun the systematic sampling of the lake Lanier microbial communities, in-situ and over time, using cutting-edge metagenomics and metatranscriptomics techniques. This work was recently funded by the NSF “Dimensions of Biodiversity” program. For further details, see:
http://www.nsf.gov/awardsearch/showAward.do?AwardNumber=124104
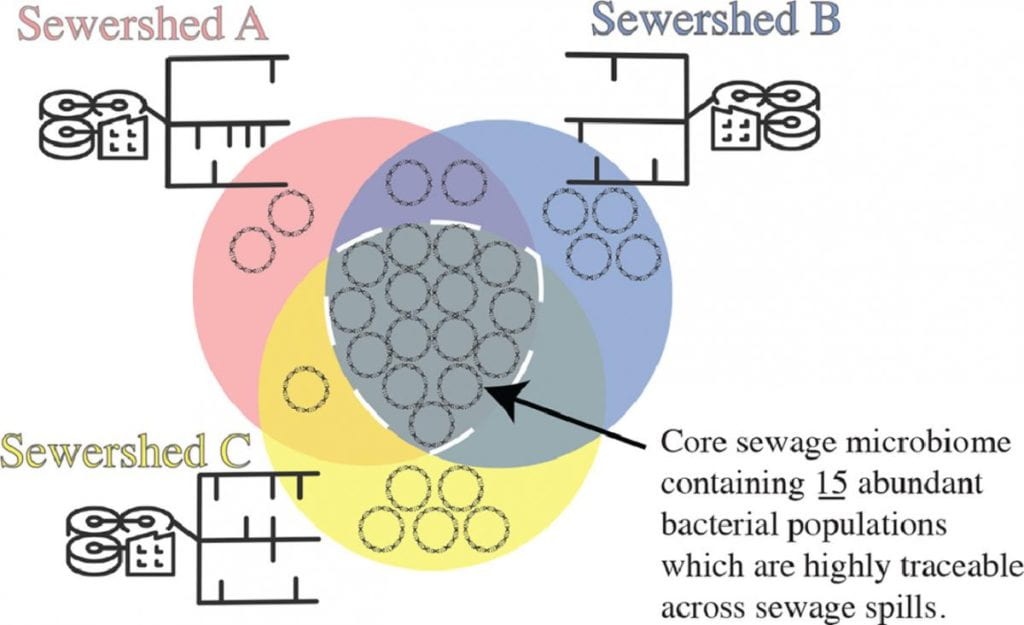
5. Microbial source tracking and fecal pollution testing
Our previous PNAS study (Luo et al., 2011), and those of others, showed that counting E. coli cells in a sample for assessing fecal or sewage contamination is frequently confounded by the presence of innocuous, environmentally adapted E. coli strains that provide a false positive signal. We are continuing this line of research and developing advanced, molecular diagnostic tests for fecal pollution testing (e.g., Lindner et al., Water Research 2021). These tests aim to robustly detect pathogenic organisms as well as genetically modified organisms via synthetic biology techniques in environmental samples and not be confounded by the co-presence of innocuous relatives. With an increasing human population on Earth, assessing water quality and the risk to human health becomes increasingly more important and an essential part of the civil infrastructure. This work was recently funded by the US EPA. For further details, see:
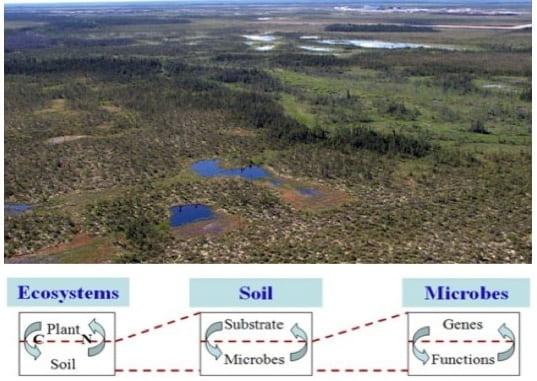
6. Microbial adaptations and N2O emissions to anthropogenic increase of CO2
With support from the US Department of Energy and NSF, and in collaboration with researchers at the University of Oklahoma, Michigan State University, University ot Tennessee and University of Illinois Urbana-Champaign, we perform comparative metagenomic analyses of microbial communities from Alaskan soils and soils of temperate and tropical (Puero Rico) regions to investigate how the corresponding microbial communities respond to increased atmospheric CO2 and temperature, especially with respect to release or sequestration of carbon and the potent greenhouse gas, N2O (nitrous oxide).
For further details, see:
7. Species interactions within dechlorinating communities
With support from the NSF and in collaboration with Dr. Loeffler (University of Tennessee), we employ a combination of microbial physiology, biochemistry and bioinformatic approaches to explore the symbiotic interactions between microbial organisms responsible for the bioremediation of important contaminants such as chlorinated hydrocarbons. For further details, see:
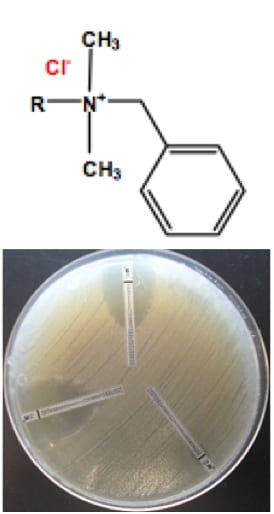
8. Controlling antibiotic resistance in engineered systems
With support from the NSF and in collaboration with Dr. Pavlostathis (Georgia Tech), we explore the role of disinfectants such as quaternary ammonia compounds in proliferating (microbial) antibiotic resistance in engineered systems, such as bioreactors of wastewater treatment plants. On a related project supported in part by CDC, we are developing metagenomic approaches and ROCker profiles to detect antibiotic resistance genes (ARGs) in environmental or clinical samples and assess the potential risk for public health. For further details, see:
http://www.nsf.gov/awardsearch/showAward.do?AwardNumber=0967130
